A typical step in the workflow for experimental sequence alignment is to trim adapter sequences, and there are many tools out there (cutadapt, AdapterRemoval, Trimmomatic, ...) for doing this. Don't use them! We think this is a bad idea, primarily because (a) it's impossible to do accurately (without the rest of the experimental context), and (b) it throws away information that can be useful in downstream alignment.
Of course, there are contexts in which adapter trimming is okay -- particularly when you are not concerned with mutations or indels, and there is no chance for ambiguity between adapter and target sequences. But such ambiguity often exists, and many experiments are in fact looking explicitly for mutations/indels.
The original SPATS pipeline -- before we inherited it -- misclassified about 1000 out of 500,000 sequences (~.2%) in a typical experiment due to these types of adapter trim issues. This will be typical of most pipelines that involve an explicit adapter trim step.
For those interested in the gory details, we include below a couple of concrete examples, both from real experiments.
A simple case, to start: the original R1 and R2 sequences are provided below, along with the adapter sequence, to be trimmed off of the right, if possible:
Since the adapter starts with A, we could potentially trim the final A off of the right end of both R1 and R2. Typically, adapter trimming tools require a longer match, so they would ignore both of these. However, when SPATS combines R1/R2 into a fragment and aligns with the other necessary targets, we get this:
SPATS knows where the target sequence should be; and based on that, it can deduce where the 4-nt handle should be. And in this case, it then knows that anything to the left of the handle indicates adapter: so it expects exactly one bp of adapter. This happens to match on both R1 and R2, so the read is matched -- fully and in context of the expectation.
Short adapter trims of this kind will always be impossible for adapter-trim tools to properly handle, as it is ambiguous whether a short match is truly adapter, or a valid part of the target sequence.
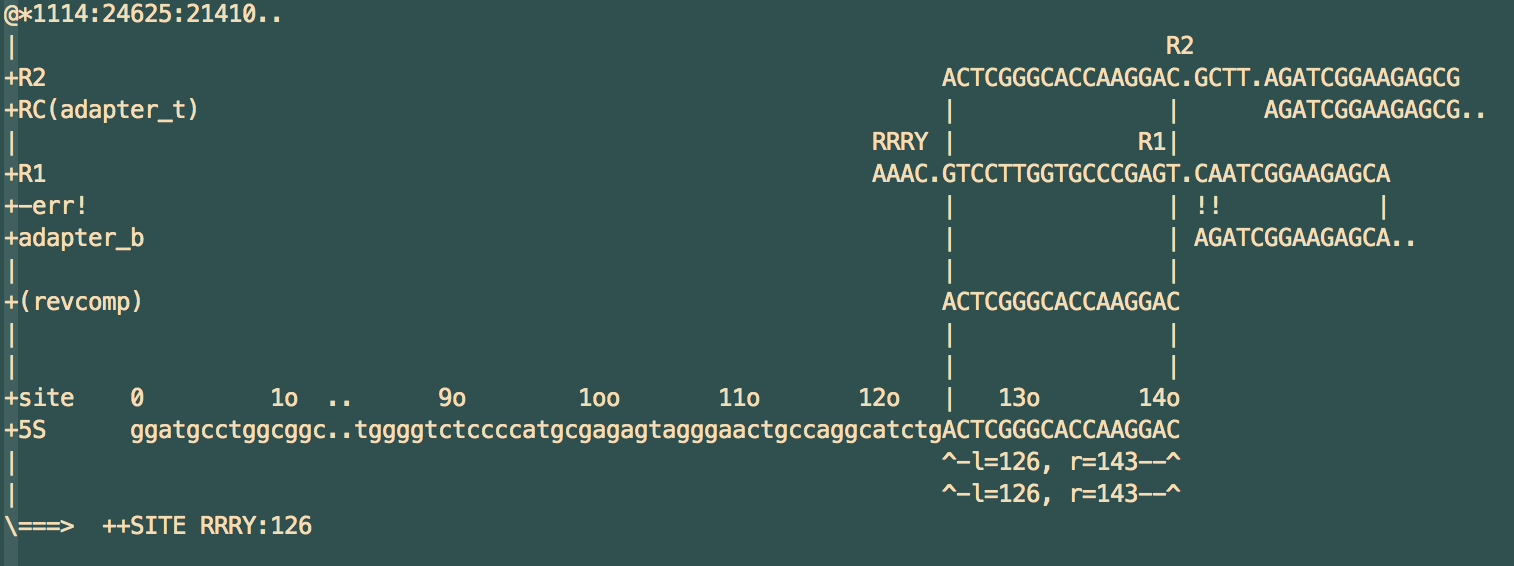
The real issue with adapter trimming, however, is due to the ambiguity inherent when dealing with mutations and indels. Consider the diagram below:
In this case, the adapter on the right side of R1 has toggle mutations in the first two bp. However, when running a typical adapter trimmer (in this case, cutadapt with default options), it leaves the C and removes AATCGG... from R1, under the assumption that a single deletion of a G is more likely than two consecutive toggles.
What's worse is that in this case, the extra C that is left, when under reverse complement, happens (25% chance!) to match the G at site 125 in the target. This caused the old version of SPATS to misclassify this pair at site 125, instead of the correct site 126.